Multiscale modeling in biology
Multiscale modeling in biology is used to take into account the complex interactions between the different organization levels in living systems. We review several models, from the most simple to the complex ones, and discuss their properties from a multiscale point of view.
Copyright Ecole de Printemps SFBT 2012
Last edited: 06/01/2013
http://sfbt-2012.imag.fr http://math.univ-lyon1.fr/~bernard/index.php?texte=SFBT
Contents
Lotka-Volterra Equations (1925)
We begin with the classic Lotka-Volterra predator-prey model
x' = x * (a - b * y)
y' = -y * (c - d * x)
The variables x and y measure the sizes of the prey and predator populations respectively. The interpretation at the population scale is the following: the growth rate of the the prey population is constant (a * x) and the death rate of the preys is proportional to the number of predators (b * y * x).
To simulate a system, we use the function called LOTKAVOLTERRA
type lotkavolterra
function xxp = lotkavolterra(~,xx)
% LOTKAVOLTERRA Predator-prey Lotka-Volterra system
%
% Examples
% sol1 = ode23(@lotkavolterra,[0 20],[ 20 20 ]);
% sol2 = ode23(@lotkavolterra,[0 20],[ 100 100 ]);
% xint = linspace(0,20,500);
% yint1 = deval(sol1,xint); yint2 = deval(sol2,xint);
% subplot(121)
% plot(xint,yint1,'LineW',1,'Color','k'); hold on
% plot(xint,yint2,'LineW',2,'Color',[0.6 0.6 0.6])
% axis square
% xlabel('time'); ylabel('population');
% subplot(122)
% plot(yint1(1,:),yint1(2,:),'LineW',1,'Color','k'); hold on
% plot(yint2(1,:),yint2(2,:),'LineW',2,'Color',[0.6 0.6 0.6])
% axis square
% xlabel('number of preys'); ylabel('number of predators');
%
% See also
%
% Ecole de Printemps SFBT 2012
x = xx(1); y = xx(2);
a = 1; b = 0.01;
c = 1; d = 0.02;
xxp = [ x * (a - b * y)
-y * (c - d * x)];
We solve the Lotka-Volterra system for times between 0 and 50, using the ode23 solver.
sol1 = ode23(@lotkavolterra,[0 20],[ 20 20 ]); sol2 = ode23(@lotkavolterra,[0 20],[ 100 100 ]);
Plot the result of the simulation two different ways.
xint = linspace(0,20,500); yint1 = deval(sol1,xint); yint2 = deval(sol2,xint); subplot(121) plot(xint,yint1,'LineW',1,'Color','k'); hold on plot(xint,yint2,'LineW',2,'Color',[0.6 0.6 0.6]) axis square xlabel('time'); ylabel('population'); subplot(122) plot(yint1(1,:),yint1(2,:),'LineW',1,'Color','k'); hold on plot(yint2(1,:),yint2(2,:),'LineW',2,'Color',[0.6 0.6 0.6]) axis square xlabel('number of preys'); ylabel('number of predators');
FKPP Equation (1937)
The Fisher-Kolmogorov-Petrovskii-Piskunov (FKPP) equation is a reaction-diffusion partial differential equation. It was introduced by R.A. Fisher in 1937 as a model for the spread of an advantageous gene in a population.
\rho_t = D \rho_{xx} + r \rho (1 - \rho)
The equation describes the frequency rho(t,x) of the advantageous gene in a population located at position x, at time t.
To simulate the equation, we use the functions fkpp and fkppbc.
type fkpp type fkppbc
function [c,f,s] = fkpp(x,t,u,DuDx)
% FKPP PDE components for the FKPP equation
%
% Examples
% x = -10:0.1:50;
% t = 0:0.1:20;
% sol = pdepe(0,@fkpp,@(x) (x<=0),@fkppbc,x,t);
% u1 = sol(:,:,1);
% imagesc(x,t,u1); axis xy;
% colormap gray
% xlabel('position x'); ylabel('time t')
%
% plot(x,u1(1,:),'LineW',2,'Color',[0.6 0.6 0.6]) % initial condition
% hold on
% plot(x,u1(21:20:201,:),'k')
% axis tight
% xlabel('position x'); ylabel('density \rho')
%
% See also: FKPPBC, PDEPE
%
% Ecole de Printemps SFBT 2012
r = 1; D = 1;
c = 1;
f = D * DuDx;
s = r * u .* (1 - u);
function [pl,ql,pr,qr] = fkppbc(xl,ul,xr,ur,t)
% FKPPBC Boundary condition for the FKPP equation
%
% See also: FKPP
%
% Ecole de Printemps SFBT 2012
pl = 0; pr = 0;
ql = 1; qr = 1;
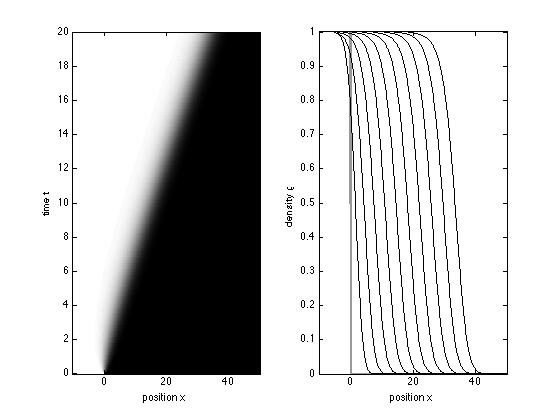
We solve the FKPP equation for times between 0 and 20 and space between -10 and 50, with an initial condition equal to one if x<=0 and zero otherwise
(x<=0)
using the PDE solve PDEPE
x = -10:0.1:50; t = 0:0.1:20; sol = pdepe(0,@fkpp,@(x) (x<=0),@fkppbc,x,t); u1 = sol(:,:,1);
Plot the result of the simulation two different ways.
figure(2) subplot(121) imagesc(x,t,u1); axis xy; colormap gray xlabel('position x'); ylabel('time t') view(2) axis tight subplot(122) plot(x,u1(1,:),'LineW',2,'Color',[0.6 0.6 0.6]) % initial condition hold on plot(x,u1(21:20:201,:),'k') axis tight xlabel('position x'); ylabel('density \rho')
Turing Equations (1952)
Alan M. Turing introduced this famous system in 1952 to show how spatial patterns (spatially heterogeneous solutions) could arise from diffusion of chemical substances. One morphogen is an activator and the other one is an inhibitor. The activator, u activates itself with a rate fu and activates the inhibitor with a rate gu. The inhibitor, v, inhibits itself with a rate gv and inhibits the activator with a rate fv. If the diffusion rate of the activator is much smaller than the diffusion rate of the inhibitor, the concentrations of u and v can show spatial instabilities.
u_t = D_u u_{xx} + fu * u + fv * v
v_t = D_v v_{xx} + gu * u + gv * v
To simulate the equations, we use the functions turing, turingbc and turingic
type turing type turingbc type turingic
function [c,f,s] = turing(x,t,u,DuDx)
% TURING PDE components for the Turing equations
%
% Examples
% x = -20:0.1:20;
% t = 0:20;
% sol = pdepe(0,@turing,@turingic,@turingbc,x,t);
% u = sol(:,:,1);
% v = sol(:,:,2);
% imagesc(x,t,u); axis xy;
% colormap gray
% xlabel('position x'); ylabel('time t')
%
% plot(x,u(1,:),'Color',[0.6 0.6 0.6],'LineW',1.5) % initial condition
% hold on
% %plot(x,v(1,:),'--','LineW',1.5) % initial condition
% plot(x,u(3:2:20,:),'k')
% %plot(x,u(3:2:20,:),'--k')
% axis tight
% xlabel('position x'); ylabel('density u')
%
% See also: TURINGIC, TURINGBC, PDEPE
%
% Ecole de Printemps SFBT 2012
fu = 0.4; fv = -0.5;
gu = 1; gv = -0.5;
f1 = fu * u(1) + fv * u(2);
f2 = gu * u(1) + gv * u(2);
Du = 0.2; Dv = 10;
c = [1 1]';
f = [Du 0; 0 Dv] * DuDx;
s = [f1 f2]';
function [pl,ql,pr,qr] = turingbc(xl,ul,xr,ur,t)
% TURINGBC Boundary conditions for Turing equations
%
% See also: TURING, TURINGIC
%
% Ecole de Printemps SFBT 2012
pl = [0 0]'; pr = [0 0]';
ql = [1 1]'; qr = [1 1]';
function u0 = turingic(x)
% TURINGIC Initial conditions for Turing equations
%
% See also: TURING, TURINGBC
%
% Ecole de Printemps SFBT 2012
u0 = [1+sin(x); 1+cos(x)];
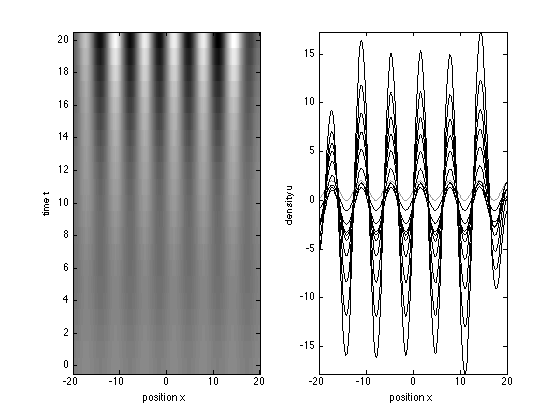
We solve the Turing equations for times between 0 and 20 and space between -20 and 20, with an initial condition given by the function turingic
x = -20:0.1:20; t = 0:20; sol = pdepe(0,@turing,@turingic,@turingbc,x,t); u = sol(:,:,1); v = sol(:,:,2);
Plot the result of the simulation two different ways.
figure(3) subplot(121) imagesc(x,t,u); axis xy; colormap gray xlabel('position x'); ylabel('time t') subplot(122) plot(x,u(1,:),'Color',[0.6 0.6 0.6],'LineW',1.5) % initial condition hold on plot(x,u(3:2:20,:),'k') axis tight xlabel('position x'); ylabel('density u')
Keller-Segel Equations (1970)
Chemotaxis is the phenomenon in which cells move according to the concentration of certain molecules in the environment. They could be attracted by food, or repelled by poisons. If the cells themselves secrete the chemotactic molecules, we can describe the movement of the cells by the celebrated Keller-Segelmodel.
u_t = d u_{xx} - (u * v)_x
v_t = epsilon * v_{xx} + u - a * v
To simulate the equations, we use the functions kellersegel , kellersegelbc and kellersegelic.
type kellersegel type kellersegelbc type kellersegelic
function [c,f,s] = kellersegel(~,~,u,DuDx)
% KELLERSEGEL PDE components for Keller-Segel equations
%
% Examples
% x = -1.5:0.01:1.5;
% t = 0:0.01:2.5;
% sol = pdepe(0,@kellersegel,@kellersegelic,@kellersegelbc,x,t);
% u = sol(:,:,1);
% v = sol(:,:,2);
% imagesc(x,t,u); axis xy;
% colormap hsv
% xlabel('position x'); ylabel('time t')
%
% plot(x,u(1,:),'Color',[0.6 0.6 0.6],'LineW',1.5) % initial condition
% hold on
% plot(x,u([26 51 101 151 201 226 251],:),'k')
% axis([-1.5 1.5 0 20])
% xlabel('position x'); ylabel('density u')
%
%
% See also: KELLERSEGELIC, KELLERSEGELBC, PDEPE
%
% Ecole de Printemps SFBT 2012
a = 0.1;
f1 = 0;
f2 = u(1) - a * u(2);
d = 0.5; epsilon = 0.05; %0.01
c = [1 1]';
f = [d, -u(1); 0 epsilon] * DuDx;
s = [f1 f2]';
function [pl,ql,pr,qr] = kellersegelbc(xl,ul,xr,ur,t)
% KELLERSEGELBC Boundary conditions for Keller-Segel equations
%
% See also: KELLERSEGEL, KELLERSEGELIC
%
% Ecole de Printemps SFBT 2012
pl = [0 0]'; pr = [0 0]';
ql = [1 1]'; qr = [1 1]';
function u0 = kellersegelic(x)
% KELLERSEGELIC Initial conditions for Keller-Segel equations
%
% See also: KELLERSEGEL, KELLERSEGELBC
%
% Ecole de Printemps SFBT 2012
c = 0.5;
d = 0.4;
u0 = [1/d*(x>=-c-d).*(x<=-c+d)+1/d*(x>=c-d).*(x<=c+d); 0];
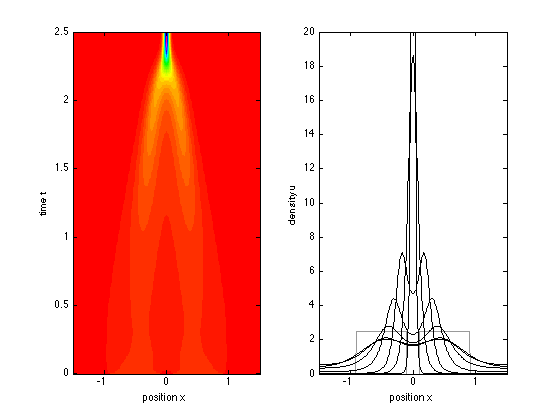
We solve the Keller-Segel equations for times between 0 and 2.5 and space between -2 and 2, with an initial condition equal to one between x = -1 and x = 1
x = -1.5:0.01:1.5; t = 0:0.01:2.5; sol = pdepe(0,@kellersegel,@kellersegelic,@kellersegelbc,x,t); u = sol(:,:,1); v = sol(:,:,2);
Plot the result of the simulation two different ways.
figure(4) subplot(121) imagesc(x,t,u); axis xy; colormap hsv xlabel('position x'); ylabel('time t') subplot(122) plot(x,u(1,:),'Color',[0.6 0.6 0.6],'LineW',1.5) % initial condition hold on plot(x,u([26 51 101 151 201 226 251],:),'k') axis([-1.5 1.5 0 20]) xlabel('position x'); ylabel('density u')
Langevin and Fokker-Planck Equations
The nonlinear Langevin equation is
y' = f(y) + g(y) * xi(t)
where xi(t) is a Gaussian, uncorrelated noise. We look at the example of the Lotka-Volterra equations
x' = x * (a - b * y) + e*xi_x
y' = -y * (c - d * x) + f*xi_y
To simulate the equations, we use the function sdeEuler
type sdeEuler
function [t,y]=sdeEuler(f,tspan,ic,sdnoise,varargin)
% SDEEULER Stochastic Differential Equation solver with the Euler method
%
% Examples
% Solve the equation y'(t) = -0.1*y(t) + 0.1*xi(t)
% with IC y(0)=1, with 10 realizations
% [t, y] = sdeEuler(@(t,y) -0.1*y, [0 10],ones(1,10),0.1*ones(1,10));
% plot(t,y,'k','LineW',0.5); axis tight
% xlabel('time t'); ylabel('y(t)')
%
% [t1, y1] = sdeEuler(@lotkavolterra, [0 60],[20 20]',3*ones(2,1));
% [t2, y2] = sdeEuler(@lotkavolterra, [0 60],[100 100]',3*ones(2,1));
% clf;
% subplot(121)
% plot(t1,y1,'LineW',1,'Color','k'); hold on
% plot(t2,y2,'LineW',1,'Color',[0.6 0.6 0.6])
% axis square
% xlabel('time'); ylabel('population');
% subplot(122)
% plot(y1(1,:),y1(2,:),'LineW',1,'Color','k'); hold on
% plot(y2(1,:),y2(2,:),'LineW',1,'Color',[0.6 0.6 0.6])
% axis square
% xlabel('number of preys'); ylabel('number of predators');
%
% Ecole de Printemps SFBT 2012
h=.01;
sh=sqrt(h);
t=tspan(1):h:tspan(2);
y=zeros(length(ic),length(t));
y(:,1)=ic;
stopcond=false;
for i=1:length(t)-1,
y(:,i+1)=y(:,i)+h*feval(f,t,y(:,i),varargin{:})+sh*sdnoise.*randn(size(sdnoise));
if stopcond && any(y(:,i+1)<0)
yz =find(y(:,i+1)<0);
y(yz,i+1)=0;
sdnoise(yz)=0;
end
end
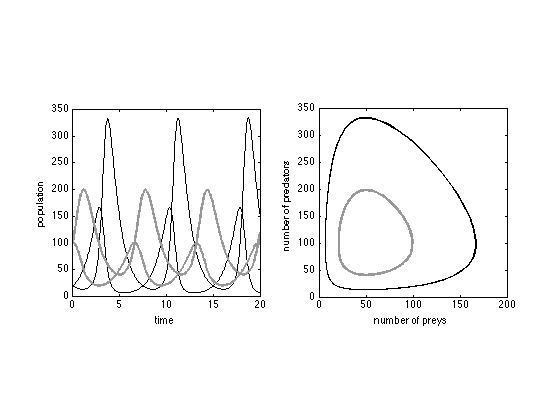
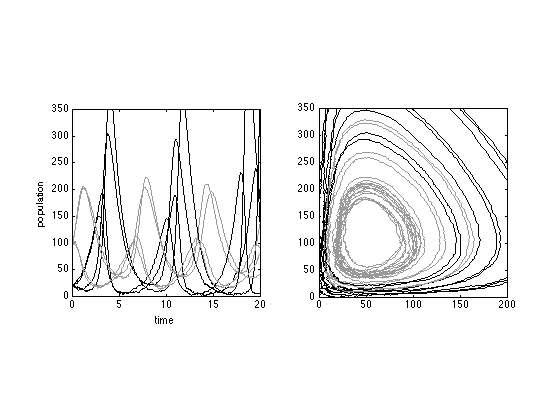
We solve the Langevin equations, with initial condition [x0,y0]'=[20 20]' and [x0,y0]'=[100 100]', with e=f=3.
[t1, y1] = sdeEuler(@lotkavolterra, [0 60],[20 20]',3*ones(2,1)); [t2, y2] = sdeEuler(@lotkavolterra, [0 60],[100 100]',3*ones(2,1));
Plot the solutions of the Langevin equation
figure(5) subplot(121) plot(t1,y1,'LineW',1,'Color','k'); hold on plot(t2,y2,'LineW',1,'Color',[0.6 0.6 0.6]) axis([0 20 0 350]) axis square xlabel('time'); ylabel('population'); subplot(122) plot(y1(1,:),y1(2,:),'LineW',1,'Color','k'); hold on plot(y2(1,:),y2(2,:),'LineW',1,'Color',[0.6 0.6 0.6]) axis([0 200 0 350]) axis square xlabel('number of preys'); ylabel('number of predators');
The Fokker-Planck equation is
p_t = - (f(y) * p)_y + 1/2 * (g(y)^2 p)_{yy}
Compare Langevin and Fokker-Planck equations. We use the functions fokkerplanck and fokkerplanckbc
type fokkerplanck type fokkerplanckbc
function [c,f,s] = fokkerplanck(x,t,u,DuDx) % FOKKERPLANCK PDE components for the Fokker-Planck equation % % Examples % x = -1:0.1:1.2; % t = 0:0.1:30; % sol = pdepe(0,@fokkerplanck,@(x) 10*(x>=1).*(x<=1.1),@fokkerplanckbc,x,t); % u1 = sol(:,:,1); % surf(x,t,u1); % % See also: FOKKERPLANCKBC, PDEPE % % Ecole de Printemps SFBT 2012 r = 0.1; D = 0.5 * (0.1)^2; c = 1; f = D * DuDx + r * x * u; s = 0; function [pl,ql,pr,qr] = fokkerplanckbc(xl,ul,xr,ur,t) % FOKKERPLANCKBC Boundary condition for the Fokker-Planck equation % % See also: FOKKERPLANCKBC % % Ecole de Printemps SFBT 2012 pl = 0; pr = 0; ql = 1; qr = 1;
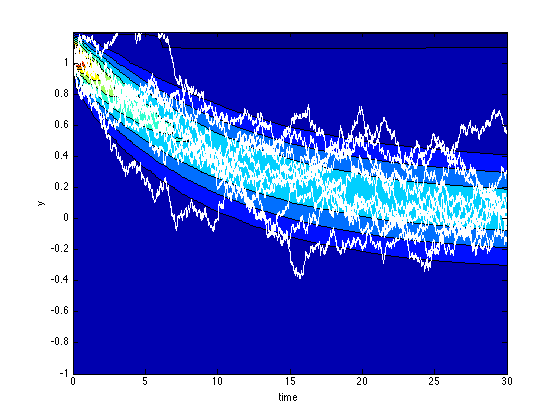
We solve the Fokker-Planck equation corresponding to the Langevin equation y' = -0.1 (y + xi), with initial conditions uniform between y=1 and y=1.1, for t=[0,30].
y = -1:0.1:1.2; t = 0:0.1:30; sol = pdepe(0,@fokkerplanck,@(y) 10*(y>=1).*(y<=1.1),@fokkerplanckbc,y,t); u1 = sol(:,:,1);
We solve the Langevin eqaution 10 times for t=[0,30], with random initial conditions distributed uniformly between y=1.0 ans y=1.1
iclangevin = rand(10,1)/10+1.0; [tl, yl] = sdeEuler(@(t,y) -0.1*y, [0 30],iclangevin,0.1*ones(10,1));
We plot and compare the solution of the Fokker-Planck equation and the solutions of the Langevin equation.
figure(6); clf contourf(t,y,u1'); hold on plot(tl,yl,'w') xlabel('time') ylabel('y')
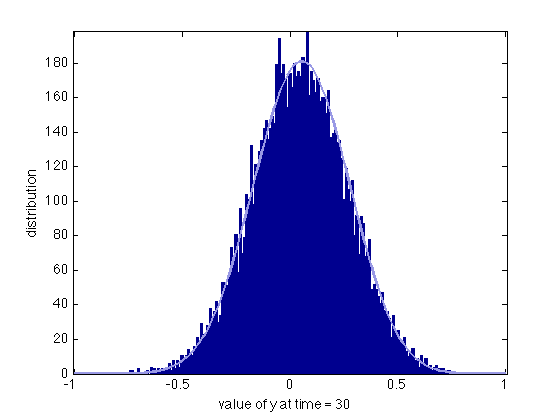
We compare the histogram of 10000 realizations of the Langevin equation at time t=30 with the solution of the Fokker-Planck equation.
runNb = 1e4; iclangevin = rand(runNb,1)/10+1.0; [tl, yl] = sdeEuler(@(t,y) -0.1*y, [0 30],iclangevin,0.1*ones(runNb,1)); figure(7); clf; binWidth = 0.01; bins = -1:binWidth:1; hist(yl(:,end),bins) hold on u1normalized = runNb*binWidth*u1(end,:)/trapz(y,u1(end,:)); u1int = interp1(y,u1normalized,bins,'spline')'; plot(bins,u1int,'LineWidth',2,'Color',[0.6 0.6 0.9]) axis tight set(gca,'FontS',14) xlabel('value of y at time = 30'); ylabel('distribution') clear y1
Hoffman et al. (2008)
Hoffmann and colleagues considered a model in which cell differentiate by moving randomly into a differentiation space.
% To simulate the equations, we use the functions hoffFK and hoffFKbc type hoffFK type hoffFKbc
function [c,f,s] = hoffFK(x,t,u,DuDx,par)
% HOFFFK PDE components for the Hoffmann et al. equation
%
% Example 1
% No differentiation, only proliferation
% ** Set r0 = 0 and r1 = 0 **
% x = [0 0.05 0.09:0.01:0.21 0.25:0.05:1];
% t = linspace(0,200,10);
% mypde = @(x,t,u,DuDx) hoffFK(x,t,u,DuDx,[0 0]);
% myic = @(x) (x>=0.1).*(x<=0.2);
% sol = pdepe(0,mypde,myic,@hoffFKbc,x,t);
% u1 = sol(:,:,1);
% surfc(x,t,u1,'LineStyle','none');
%
% Example 2
% Differentiation and proliferation
% ** Set r0 = 0.01 and r1 = 0.5 **
% x = 0:0.1:1;
% t = linspace(0,200,10);
% mypde = @(x,t,u,DuDx) hoffFK(x,t,u,DuDx,[0.01 0.05]);
% myic = @(x) (x>=0.1).*(x<=0.2);
% sol = pdepe(0,mypde,myic,@hoffFKbc,x,t);
% u1 = sol(:,:,1);
% surfc(x,t,u1);
%
% See also: HOFFFKBC, PDEPE
%
% Ecole de Printemps SFBT 2012
maxProlif = 0.02;
r0 = par(1);
r1 = par(2);
maxNoise = 0.2;
minNoise = 0.1;
c = 1;
f = 0.5*(differentiation(x).*sigma2(x).*DuDx + ...
(differentiation(x).*ds2(x)+ddr(x).*sigma2(x)).*u);
s = prolif(x)*u;
function y = prolif(x)
% PROLIF is the proliferation rate of x-cells
y = 4*maxProlif*x.*(1-x);
end
function y = differentiation(x)
% DIFFERENTIATION is the differentiation rate of x-cells
y = r0 + r1*prolif(x);
end
function y = ddr(x)
% DDR derivative of DIFFERENTIATION
% Used to compute f
y = r1*4*maxProlif*(1-2*x);
end
function y = sigma2(x)
% SIGMA2 differentiation noise
y = minNoise + 4*maxNoise*(x-0.5).^2;
end
function y = ds2(x)
% DS2 derivative of SIGMA2
% Used to compute f
y = 8*maxNoise*(x-0.5);
end
end
function [pl,ql,pr,qr] = hoffFKbc(xl,ul,xr,ur,t)
%HOFFFKBC Boundary condition for the Hoffmann et al. equation
%
% See also: HOFFFK
%
% Ecole de Printemps SFBT 2012
pl = 0; pr = 0;
ql = 1; qr = 1;
We solve the Hoffmann equation for t=[0,200] and space x=[0,1], with an initial condition equal to one for x=[0.1,0.2].
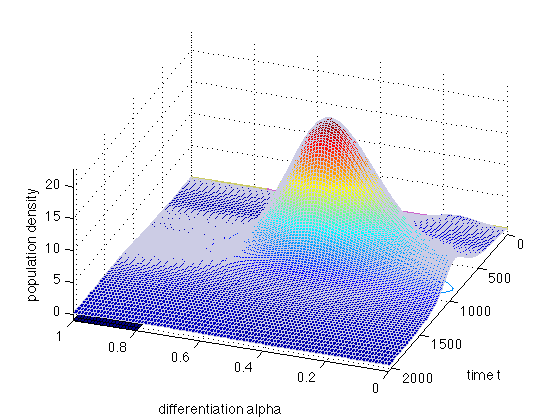
In the first example, we solve the equation when there is no differentiation: R(alpha)=0. Set r0=0.0 and r1=0.0.
x = [0 0.05 0.09:0.01:0.21 0.25:0.05:1]; t = linspace(0,200,10); mypde = @(x,t,u,DuDx) hoffFK(x,t,u,DuDx,[0 0]); myic = @(x) (x>=0.1).*(x<=0.2); sol = pdepe(0,mypde,myic,@hoffFKbc,x,t); u1 = sol(:,:,1); figure(8); clf; surfc(x,t,u1,'EdgeColor',[0.4 0.4 0.4]); set(gca,'FontS',14); xlabel('differentiation alpha'); ylabel('time t'); zlabel('population density') view(-159.5,44); hold on plot3(x,0*x,4*8*x.*(1-x),'LineWidth',2,'Color',[0.8 0.4 0.4])
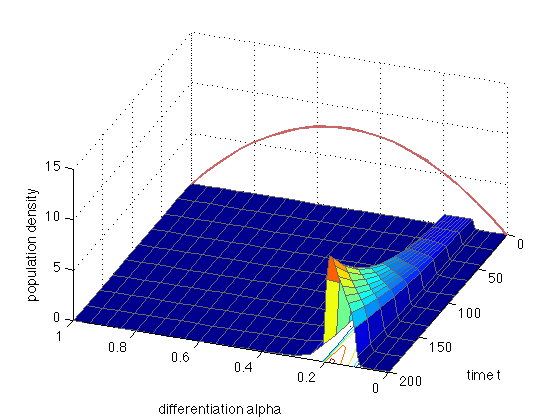
The red curve is the proliferation rate r(alpha) (in arbitrary units).
In the second example, we solve the equation when there is differentiation: R(alpha)=r0+r1*r(alpha). Set r0 = 0.01 and r1= 0.05.
x = 0:0.1:1; t = linspace(0,200,10); mypde = @(x,t,u,DuDx) hoffFK(x,t,u,DuDx,[0.01 0.05]); myic = @(x) (x>=0.1).*(x<=0.2); sol = pdepe(0,mypde,myic,@hoffFKbc,x,t); u1 = sol(:,:,1); figure(9); clf; surfc(x,t,u1,'EdgeColor',[0.4 0.4 0.4]); set(gca,'FontS',14); xlabel('differentiation alpha'); ylabel('time t'); zlabel('population density') view(-159.5,44); hold on plot3(x,0*x,4*4*x.*(1-x),'LineWidth',2,'Color',[0.8 0.4 0.4]) plot3(x,0*x,0.01+0.5*4*4*x.*(1-x),'LineWidth',2,'Color',[0.8 0.4 0.8]) plot3(x,0*x,0.1 + 4*3*(x-0.5).^2,'LineWidth',2,'Color',[0.8 0.8 0.4])
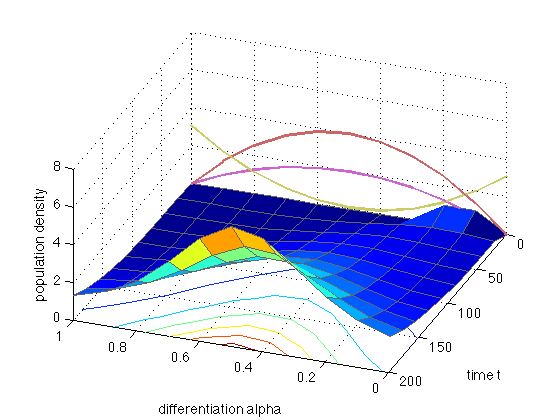
The red curve is the proliferation rate r(alpha). The green curve is the noise sigma2(alpha) and the blue curve is the differentiation rate R(alpha) (all in arbitrary units).
In the third example, we solve the equation with a growth limited by the total population.
% To simulate the equations, we use the function |mscelldiff| help mscelldiff
MSCELLDIFF Nonlinear PDE with population-dependent growth
MSCELLDIFF solve the modified Fokker-Planck equation adapted from
Hoffmann et al. (2008). The PDE is
[1] * D_ [u] = D_ [0.5*(R*s2*DuDx+(R*ds2+dR*s2)*u)] + [r(x)*u*(1-NA/K)]
Dt Dx
---- --- --------------------------------- ---------
c u f(x,t,u,Du/Dx) s(x,t,u,Du/Dx)
where
x differentiation level x in [0,1]
t time
u = u(t,x) cell density at differentiation level x
R = R(x) differentiation rate
dR = dR(x) derivative of the differentiation rate
s2 = s2(x) variance of the differentiation jumps
ds2= ds2(x) derivative of s2
r = r(x) proliferation rate
NA = cell population acting on the feedback
K = carrying capacity
Ecole de Printemps SFBT 2012
Published output in the Help browser
showdemo mscelldiff
We run the system with default parameters
figure(10); clf; mscelldiff;
-------------------------------------------------- *******************************************